More About the Job.
Beginning bacteriology lab courses generally confine themselves to the relatively small universe of conveniently easy-to-grow representatives of the chemoorganotrophic bacteria. Much can be learned – and applied to other living things – from this microbial group which does happen to include a lot of important pathogens, food spoilage organisms, general contaminants, and participants in various aspects of simple biodegradation – basically the "in our face" organisms. As in our previous "Bacteriology 102" course, the new, partly-virtualized Microbiology 102 course encompasses a lot of good things to know about (including anaerobic respiration), and (until I retired) it continued to venture beyond chemotrophy into the wonderful world of anoxygenic phototrophy. Indeed, we found that the purple non-sulfur photosynthetic bacteria are a refreshing change of pace and are actually quite easy to isolate, even from hailstones!
A beginning lab course (such as ours) should really be spending some time in the fascinating worlds of cyanobacteria and chemolithotrophs. That these organisms tend to be "inconvenient" to work with is no reason to ignore them. Think about where we would be without them. So why just pay lip service to microbial diversity? In the 21st Century we may discover presently unimaginable metabolic processes that energize and fabricate life forms on Mars and elsewhere, and putting more emphasis right now on lithotrophic organisms in general microbiology teaching labs would not be a bad idea. Such has always been highly relevant to microbiology. The late P. W. Wilson made the study of the nitrogen cycle (including nitrification – an important chemolithotrophic process in the soil) a significant part of his introductory micro lab when he was here through the early 1970s.
 |
 |
Certainly microbial consortia are a practical consideration for demonstration and discussion. In my explorations of sandy areas up north, I frequently come across classic examples of cryptobiotic soil – those places where mixtures of chemotrophic and phototrophic microorganisms turn the inorganic into organic and initiate substrates for higher forms of life. These organisms are Nature's Terraformers – important in the early stages of the process wherein sand develops into topsoil. They can appear as clumps of steel wool and as velvety pincushions, and cutting one open reveals a core of sand. Click on the two nearby photos which were taken while visiting the Apostle Islands where there are some restricted areas containing cryptobiotic soil that are marked off as delicate ecosystems. Microscopically one can readily see fungi and also filamentous and unicellular algae and cyanobacteria. I have yet to check out what bacteria may be present. Aside from these "soil lichens," iron bacteria are another mixed group of organisms that I have been treating superficially (so far) on the web.

Of the (at least) five general ways that organisms can generate their energy, we have delved into all but oxygenic phototrophy in Microbiology 102. Along the way we realize that testing and describing organisms regarding their use of any one (or more) of these five catabolic methods is certainly more instructive than getting bogged down with the old-school method (with the Thioglycollate Medium deeps) of determining "oxygen relationships" which can only be applied productively to the characterization of those aforementioned easy-to-grow chemoorganotrophs and is often a pain for students and instructors to interpret. Students can get the false idea that all organisms can be characterized by this method and that the chemical nature of the medium (i.e., how thioglycollate and cysteine reduce the environment) is more important than why the glucose is essential. It is an expensive, redundant and generally ill-taught test which is better used to demonstrate the special growth patterns of microaerophiles and the relative efficiencies of fermentation vs. aerobic respiration as shown for those named in this test system as "facultative anaerobes." But instructors happily stay mired in their 20th century habits, explaining an organism's growth pattern as whether or not it "likes" oxygen – disdaining catabolic reasons and ultimately wasting a lot of money.
How is this test redundant? Easy, when you consider the actual reasons for the growth patterns in Thioglycollate Medium and what alternative, inexpensive and easier-to-read tests can indicate – namely, whether the organism can (1) tolerate a normal aerobic environment, (2) respire aerobically and (3) ferment. The catalase test correlates with aerobic respiration, and fermentation (with the resulting anaerobic growth) can be tested in Glucose Fermentation Broth.
Many organisms have other reasons why they can grow aerobically or anaerobically, and most species of bacteria in this world can't even grow in our usual laboratory media anyway.
Examples of how to be instructive and expansive without the use of the usual oxygen relationship test:
- E. coli should be thought of as an organism that does aerobic respiration, anaerobic respiration and fermentation as tested with the catalase, nitrate reduction and glucose fermentation tests. Anaerobic growth goes along with the positive reactions in the latter two tests. (By the way, if you leave glucose out of Thioglycollate Medium, E. coli will appear as a strict aerobe.)
- We can test purple non-sulfur photosynthetic bacteria and see that they can grow aerobically by aerobic respiration and anaerobically by anoxygenic phototrophy. How we do this in Microbiology 102 is shown here. (And if we were to test these organisms with Thioglycollate Medium, they would only appear as strict aerobes. Except for some rare strains, these organisms would not be able to grow anaerobically as they do not ferment.)
Even though this test should have died out with the 20th Century or at least with the demolition of Fred Hall, I actually have a suprisingly objective treatment of the traditional "oxygen relationship test" which explains its limited usefulness here. I miss the real good old days back in 5th grade science class when "oxygen relationships" were simply described as whether or not an organism had the capability (for whatever reason) of growing aerobically and/or anaerobically – with the simple use of the terms "aerobe," "anaerobe" and "facultative" [sic]. (Facultative, like preventive, is only a modifier. Preventative, by the way, should only be used as a noun.)

 I tend to spend an awful lot of time trying to find less time-consuming ways to teach the more complicated stuff, and there is no time to get cute and condescending in presenting the subject matter. After I can overcome the frequent mental blocks and get things understandable, then the oral presentations in lab become more effectual, and the board diagrams, handouts, manual material and associated web pages write themselves. I tend to spend an awful lot of time trying to find less time-consuming ways to teach the more complicated stuff, and there is no time to get cute and condescending in presenting the subject matter. After I can overcome the frequent mental blocks and get things understandable, then the oral presentations in lab become more effectual, and the board diagrams, handouts, manual material and associated web pages write themselves.
Regarding the webworks, I am still pumping out the not-so-interactive reference material in an attempt to upload what little I know about bacteriology besides what is in the 2006 lab manual cited below. The following apples/oranges list reflects some points:
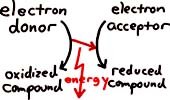
- My original catabolism page (still ranking reasonably high in a Google search for "catabolism" but no longer number one) arose out of an attempt to summarize energy/electron/ATP generation in the Farm Microbiology Short Course where lecture time was very short indeed. Farm Microbiology was a bright spot in the school year and certainly a place to expand on soil microbiology and lithotrophy. Summaries of microbiology-related concepts which were developed in this course helped considerably in teaching other courses. Eventually a couple handouts – nos. 1 and 2 – resulted to summarize catabolic procedures in enough detail for the current Microbiology 102 course. Illustrated in these handouts are the correct definitions and differentiations of the terms lithotroph vs. organotroph and also chemotroph vs. phototroph – keeping in mind respiration and fermentation only apply to the chemotrophic organisms.
- It is always best to summarize the basic strategy of any biological process first. For catabolism, then, further details can come along as time and relevancy to the course permit and may include such things as specific intermediates and pathways, the internal electron transfers in fermentation pathways, the importance of NADP as an electron acceptor in phototrophy, cyclic vs. non-cyclic photophosphorylation, what photoorganotrophy really amounts to, and the interesting variations of anaerobic respiration presented by methanogenesis and "anammox." (Anammox should be included in any discussion of the nitrogen cycle.)
- Another web effort is the beta-galactosidase page which is hidden away here.
- More bacteriological concepts whose practical applications and interpretations tend to follow basic patterns include quantitation, media, isolation, identification, and the cycles of elements. These things – as well as proper usage of terms – are gone over in the web pages expanded upon below and also in the 2006 manual (also below).
- The enteric bacteria are especially suited as practical examples to use in the discussions regarding differential media formulation (see references 3 and 4 below) and genotypic identification, and that is where I tend to concentrate my interest concerning these organisms. When dealing with enterics in the teaching lab over the past several decades, useless memorization of genera and species and their characteristics and habitats have long ago given way to a more practical approach. A handout associated with our enteric experiment (which includes one of our classic "thought questions") is shown here.
The above-mentioned web material is simply what I have let happen, and I have no intention of making work for myself (and thereby creating misery) by putting together something more all-inclusive. Enough qualified individuals are posting on-line textbooks, and my interests in microbiology may not be all that comprehensive. Doing the web thing works best for me when I can go at my own speed. Thank you very much.
One kind of writing course that should be required concerns something I entirely regret not getting the hang of decades ago, and that is taking lecture notes in real-time that are complete, organized, and understandable. Maybe I should have learned stenography in high school. The inability to generate a "things to do" list that is totally inspiring and easy to adhere to has been another problem. Yet when it comes to writing in general – whether it be academic, travel-related, fiction or whatever – who cannot be inspired by the intricate but clearly expressed writing of such prolific authors as Leslie Arlen.
Getting back to topic, here is a generality that seems to hold up well: All dilution plating problems are basically the same problem but with different variables. That same idea applies when one is interpreting the various pH-based differential media. The solutions to these things need not be made unnecessarily complicated, and each particular medium or dilution problem need not be dealt with from scratch as a special case, for crying out loud.
No lab protocol is perfect in its content or organization, and there is always a need for improving such things. Students can look at a manual protocol for an upcoming experiment and come up with a clarified flow chart to get the lab done efficiently. Also, with a good command of basic microbiology, any thoughtful instructor or student can come up with some really creative ways of doing and finding out certain things, and it is encouragement of that sort of thing that is behind the thought exercise here.

Some things I will not go along with:
- Would you believe there are still those individuals who teach their students that an autotroph is defined as one that "makes its own food"? No organism can do that for cryin' out loud, as every living thing rearranges its incoming nutrients (organic and inorganic) for catabolism and biosynthesis according to its kind. If "food" is meant to imply organic nutrients ("made" from carbon dioxide), then say so!
- The expanding redefinition of "mentor" (and "mentoring," etc.) may seem to obliterate the original meaning of the term which suggests a helpful personal and/or professional relationship that is simply allowed to happen – and for free! One might wonder how that can be mandated by some specially-funded program which is basically advising? At any rate, I'm starting to lighten up about this. And here is a positive note: I have seen random acts of genuine mentorism during my decades here in the Department of Bacteriology, and such has been quite uplifting for all concerned.
- "Aliquot" has unfortunately come to mean any specified volume rather than its originally intended meaning which still has practical application wherein a better word cannot substitute.
- "Strain" is a commonly mis-used word which can get so overly-used that it sounds ugly. (The same applies to the above-named terms.) Clarification is hopefully made here, and I stand by that discussion 100%.
- "Coliform" is not a taxonomic subgroup of the "enteric" organisms. All too often one sees a list of genera (Escherichia, Klebsiella, etc.) "classified" as such, even though many of their strains do not ferment lactose at all. It so happens that the vast majority of coliform isolates are ultimately identified as a member of the enteric family, but that certainly does not mean coliforms should be necessarily defined as "enterics." More about coliforms that I have been teaching is here.
- As for is, it is becoming the fashion to say "media is" and "bacteria is" even among grown PhD-types who actually speak that way in public. The real singular forms of these nouns seem to be disappearing.
- How can an instructor say "lactose is the carbon source" in the explanation of lactose fermentation which is a catabolic process – not anabolic!
- And then students somehow gain the impression that the abbreviation "i.e." means "for example" which is an impossible stretch as explained here.
As one structures one's course over its term and expects it to be a firm foundation for further coursework and productive research, "mixing apples and oranges" and otherwise playing loose with the basic concepts and definitions will turn it into a house of cards. Furthermore, graduating ignorant and spending one's professional life addicted to consultants is not part of anyone's definition of "leadership."
I do what I can to serve the subject matter as correctly and as well-organized as I can. Microbiology 102 is not meant to be taught by relative amateurs.

Working with the teaching labs' culture collection over the several decades of my employment here has been quite educational. A few of our stock cultures are descendants of ATCC strains (many generations removed), but the majority of our cultures have been isolated by myself or students and have wound up being "archived" for one reason or another. My "ET Project" introduced a number of enterics into our teaching labs including a strain of the very rare Edwardsiella tarda Biogroup 1 whose origin was a small, warm bay of Lake Superior which harbored a lot of seagull feces and dead fish. Biogroup 1 of E. tarda is sometimes mentioned in the literature as being inherently H2S-negative. However, in some media such as TSI Agar, detection of the actual H2S production can be negated by excess acid production from lactose and/or sucrose fermentation. Biogroup 1 shows a good H2S-positive reaction in other media such as KIA and the API-20E test.
Our spectacular Photobacterium was isolated from store-bought shrimp in the early 1980s by our "Advanced General Bacteriology" lab course (the late and lamented Bacteriology 320). The strains of E. coli and K. pneumoniae that we use routinely were isolated from pitcher plants, and our Rhodomicrobium came from water trapped by a bromeliad; click here for more about these plant sources.
Even though some of our cultures come from well-known, commercial sources, we cannot put any strain number on any of our cultures but our own. Stock culture curators may not be able to prove that any particular strain of theirs has the same exact genotype and phenotype of the isolate or certified strain it was descended from – no matter how few generations have transpired. For certified cultures for research or industry, one must go to suppliers of such cultures – for example, ARS/NRRL and ATCC.
Lyophilization is probably the best method of storing cultures for indefinite periods, and it was an ongoing process when our 1940's-era machine could be MacGyvered to work reliably. Lately I have come across a storage method that involves methylcellulose in which one does not have to put up with problems associated with lyophilization or ultra-low temperature freezing. Testing the reliability of this method is pending.
We have experienced the loss of some cultures while in refrigerated storage. One lesson I have learned is never to store Edwardsiella tarda on Heart Infusion Agar. Nutrient Agar works better for extended viability under refrigeration (six months; perhaps more) for reasons unknown.
Also, we have seen major characteristics of a few of our refrigerator-stored strains visibly shift during serial subculturing. Some examples follow:
- The type strain of Pseudomonas aeruginosa loses its intense blue pigment upon repeated transfer.
- A Streptomyces griseus strain we used as a good "positive control" for showing filamentous growth and antibiotic production lost both properties in a few years. We now use an unspeciated student isolate that continues to be reliably spectacular on both accounts.
- An occasional mutant in our Klebsiella pneumoniae strain that produces no capsule (and consequently an atypical colony on EMB and MacConkey Agars) can show itself in great numbers after a few serial subcultures – necessitating the re-isolation of a typical mucoid colony before using the culture in class.
- A minority component (probably a mutant) in many of my Edwardsiella tarda cultures which has a delayed hydrogen sulfide reaction can "take over" after very few serial transfers of the cultures! This has happened along with a decrease in alkaligenic activity (less amino acid deamination and/or lysine decarboxylation) – resulting in colonies (tested on our Edwardsiella isolation medium mentioned in Reference 3 below) that give a net acidic reaction instead of the expected alkaline reaction. Come to think of it, the greater acidity may be the factor behind the decreased detection of hydrogen sulfide (as can happen in TSI Agar).
So, what is frequently a tedious and thankless chore can also be a kind of education one cannot pick up from one's usual coursework or laboratory teaching duties.
- Robert H. Deibel and John A. Lindquist. 1981. General Food Microbiology Laboratory Manual. Pearson Education, Paramus, NJ. ISBN 0-8087-5559-5. In this lab manual, the real principles of food microbiology are dealt with in organized fashion and include: (1) detection and identification of contaminants, spoilage organisms, food-borne pathogens, and indicator organisms; (2) production of fermented foods by wild fermentation and with the aid of starter cultures; (3) respect for and practice of the concepts of aseptic technique in the laboratory and compartmentalization in food processing; and (4) some experiments involving microbial growth and the control of such growth. Regarding the last point, an understanding of the concept of "water activity" is most important. In the giddy rush to modernize courses, a lot of basic things tend to get discarded. (Throw out baby; keep bathwater.) However, these are items that should be part of any food microbiology lab course as is a discussion of the art and science of epidemiology as it relates to tracing the course of an outbreak of foodborne disease. Common sense from a real food microbiologist (R.H.D.) infuses each page of this manual, and my usual blanket (even though benign) disparagement of lab manuals does not apply here.
The bacteriological nomenclature may be a bit dated, but at least by 1981 we finally got away from using "Aerobacter." However, that genus name seems to have made a virtual comeback now that the classic "Aerobacter aerogenes" strains – which have been lately classified as Enterobacter aerogenes (motile) and Klebsiella pneumoniae (non-motile) – are closely allied again with the creation of Klebsiella mobilis for the motile strains. (Enterobacter aerogenes is now synonymous with Klebsiella mobilis in the new Bergey's Manual which has a citation below.)
- John A. Lindquist. 1975. Bacteriological and Ecological Observations on the Northern Pitcher Plant, Sarracenia purpurea. M.S. Thesis, Department of Bacteriology, University of Wisconsin, Madison, WI. This project came about when my proposal to pursue a coherent problem involving cyanophages was summarily rejected. My question was this: Is lysogenic conversion responsible for Microcystis aeruginosa toxicity?
So, while writing the thesis back in the 1970s, I was mentioning that my work was only "preliminary" – not explaining well enough that I knew of others who could likely use it as a reference in their research on the subject, as I would have better ideas to pursue if I ever intended to go on for the Ph.D. at some later time. Incidentally, information in my thesis turned out to be quite helpful to carnivorous plant researchers; so that mission was accomplished. However, back in the '70s, the powers that be took "preliminary" to mean that I intended to continue on the subject of pitcher plants for the Ph.D. and said no to my going beyond the masters level, then and there. Following my real intentions, I was just happy to stick with the ongoing full-time job as lab instructor which I think turned out quite well. Thank you very much.
Recently, after a couple of decades away from reading any of my thesis, I came away from it feeling like it really did have a lot of actual microbiological substance. One should expect to find the proteolytic and chitinolytic bacteria useful in breaking down insects, but who would have thought these plants were such a repository of E. coli and unusual organisms such as the purple non-sulfur photosynthetic bacteria? So, before getting excited and e-mailing me about carnivorous plants in general, please go to this page first and check out the disclaimers.
When it comes to green and leafy life-forms, I have been finding mulleins to be quite fascinating and instructive. Maybe they will find some practical use above and beyond their alleged medicinal value. Considering the abundance of cellulose in these ubiquitous plants, perhaps mulleins can contribute to the production of cellulosic ethanol? Or maybe not. When it comes to providing for our own energy needs from whatever source we may possess in abundance, we have been stalled most miserably.
 John A. Lindquist. 1991. Medium and Procedure for the Direct, Selective Isolation of Edwardsiella tarda from Environmental Water Samples. (Poster presented at ASM Meeting in Dallas on May 8, 1991.) Abstr. Annu. Meet. Am. Soc. Microbiol. 1991, C-303, p. 302. This project showed how a hypothetical selective-differential medium to isolate a specific physiological type of bacterium can work for real as it does on paper. Even in the 21st Century, this sort of thing still has relevance in the isolation and culture of pathogens and other organisms of interest from the environment. More about the "programming" of such media is here. This truly is great fun and is highly instructive – easily fitting in with lab teaching. John A. Lindquist. 1991. Medium and Procedure for the Direct, Selective Isolation of Edwardsiella tarda from Environmental Water Samples. (Poster presented at ASM Meeting in Dallas on May 8, 1991.) Abstr. Annu. Meet. Am. Soc. Microbiol. 1991, C-303, p. 302. This project showed how a hypothetical selective-differential medium to isolate a specific physiological type of bacterium can work for real as it does on paper. Even in the 21st Century, this sort of thing still has relevance in the isolation and culture of pathogens and other organisms of interest from the environment. More about the "programming" of such media is here. This truly is great fun and is highly instructive – easily fitting in with lab teaching.
My long-awaited (?) web page explaining the use of "ET Agar" in the isolation of Edwardsiella tarda is finally on the web here, and the medium is discussed briefly in the Edwardsiella chapter of The Prokaryotes.
Not fun was another project involving searching for a gram-positive pathogen among soil samples, but the isolation medium that came out of that made the whole ordeal worthwhile.
 John A. Lindquist and J. J. Farmer III. 1999. Isolation and Characterization of a New Genus of Edwardsiella-like Bacteria from Wisconsin Lakes. (Poster presented at ASM Meeting in Chicago on May 31, 1999.) Abstr. Annu. Meet. Am. Soc. Microbiol. 1999, R-1, p. 616. This is something brand new that tried to pass itself off as Edwardsiella tarda on the isolation medium mentioned in the preceding reference. After consulting the enteric identification tables in recent editions of the Manual of Clinical Microbiology (ISBN 1-55581-371-2), it appears that this organism is distinctive among the gram-negative, oxidase-negative fermenting rods in being both arginine-positive and mannitol-negative. With this in mind, a selective-differential medium for this organism was formulated and used successfully. The Enteric Identification Lab of the CDC did a complete workup of the organism's phenotypic and genotypic characterization, and it has been established as CDC Enteric Group 121. GenBank has its 16S rRNA sequence on the web: Click here, choose Nucleotide in the drop-down menu, and search for AF015258. The tentative name of the organism that you will see there translates as "water unit from Hayward"! The pond where I found the first isolate is shown here. This small pond was originally dug out of the sphagnum swamp by my grandfather about fifty years earlier. Why the manuscript containing the complete story from isolation through characterization remains sequestered at the CDC is beyond me. John A. Lindquist and J. J. Farmer III. 1999. Isolation and Characterization of a New Genus of Edwardsiella-like Bacteria from Wisconsin Lakes. (Poster presented at ASM Meeting in Chicago on May 31, 1999.) Abstr. Annu. Meet. Am. Soc. Microbiol. 1999, R-1, p. 616. This is something brand new that tried to pass itself off as Edwardsiella tarda on the isolation medium mentioned in the preceding reference. After consulting the enteric identification tables in recent editions of the Manual of Clinical Microbiology (ISBN 1-55581-371-2), it appears that this organism is distinctive among the gram-negative, oxidase-negative fermenting rods in being both arginine-positive and mannitol-negative. With this in mind, a selective-differential medium for this organism was formulated and used successfully. The Enteric Identification Lab of the CDC did a complete workup of the organism's phenotypic and genotypic characterization, and it has been established as CDC Enteric Group 121. GenBank has its 16S rRNA sequence on the web: Click here, choose Nucleotide in the drop-down menu, and search for AF015258. The tentative name of the organism that you will see there translates as "water unit from Hayward"! The pond where I found the first isolate is shown here. This small pond was originally dug out of the sphagnum swamp by my grandfather about fifty years earlier. Why the manuscript containing the complete story from isolation through characterization remains sequestered at the CDC is beyond me.
- John A. Lindquist and J. J. Farmer III. 2005. Genus XXXVIII. Trabulsiella. In Brenner, Krieg and Staley (eds.), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, pp. 827-828. Springer, New York. ISBN 0-387-24144-2. No, this isn't the new genus mentioned just above. Stay tuned for that!
- John A. Lindquist. 2006. General Microbiology: A Laboratory Manual, Fourth Edition. McGraw-Hill Companies. ISBN 0-07-339101-8. This is the edition of the manual that we used during the last semester that Bacteriology 102 was taught as an entirely hands-on lab course. Over its last decade, the course and its manual were supplemented by the first two websites mentioned in No. 8, below.
 Henry S. Gibbons, Stacey M. Broomall, Lauren A. McNew, Hajnalka Daligault, Carol Chapman, David Bruce, Mark Karavis, Michael Krepps, Paul A. McGregor, Charles Hong, Kyong H. Park, Arya Akmal, Andrew Feldman, Jeffrey S. Lin, Wenling E. Chang, Brandon W. Higgs, Plamen Demirev, John Lindquist, Alvin Liem, Ed Fochler, Timothy D. Read, Roxanne Tapia, Shannon Johnson, Kimberly A. Bishop-Lilly, Chris Detter, Cliff Han, Shanmuga Shozhamannan, C. Nicole Rosenzweig, Evan W. Skowronski. 2011. Genomic Signatures of Strain Selection and Enhancement in Bacillus atrophaeus var. globigii, a Historical Biowarfare Simulant. PLoS ONE 6(3): e17836. doi:10.1371/journal.pone.0017836 This article is found on-line here. Henry S. Gibbons, Stacey M. Broomall, Lauren A. McNew, Hajnalka Daligault, Carol Chapman, David Bruce, Mark Karavis, Michael Krepps, Paul A. McGregor, Charles Hong, Kyong H. Park, Arya Akmal, Andrew Feldman, Jeffrey S. Lin, Wenling E. Chang, Brandon W. Higgs, Plamen Demirev, John Lindquist, Alvin Liem, Ed Fochler, Timothy D. Read, Roxanne Tapia, Shannon Johnson, Kimberly A. Bishop-Lilly, Chris Detter, Cliff Han, Shanmuga Shozhamannan, C. Nicole Rosenzweig, Evan W. Skowronski. 2011. Genomic Signatures of Strain Selection and Enhancement in Bacillus atrophaeus var. globigii, a Historical Biowarfare Simulant. PLoS ONE 6(3): e17836. doi:10.1371/journal.pone.0017836 This article is found on-line here.
-
 Since 1997, I have been putting bacteriology-related material on the web to help with teaching various laboratory courses. Handouts to go along with the lectures in these courses have contributed to – and have been derived from – much of this material: Since 1997, I have been putting bacteriology-related material on the web to help with teaching various laboratory courses. Handouts to go along with the lectures in these courses have contributed to – and have been derived from – much of this material:
- The "GENERAL MICRO LAB TOPICS WEB PAGES" are now on jlindquist.com and are indexed here. Most had their origins in review or remedial material provided as handouts when I taught the Bacteriology/Food Science 324 lab course way back in the last millennium. One is expected to have a good general microbiological background and understand this material fully when dealing with (1) federal and industry manuals which detail methods and regulations and are not meant to serve as textbooks, (2) reference works such as Bergey's Manual and The Prokaryotes, and (3) practicing microbiologists out there in the real world who (by the way) occasionally express serious concern about new hires who have not retained their proper aseptic technique procedures since their graduation.
- The "semi-retired" BACTERIOLOGY 102 SITE is on splammo.net and was freshened-up for each semester that I happily taught 102 as a totally hands-on laboratory course. (The impetus for this site grew out of the thought that every course taught at a publicly-funded university should have a publicly-accessible site on the internet.) This went on until the course became partially virtualized in the Spring Semester of 2007 along with becoming re-named "Microbiology 102." The last iteration of this website begins at its home page and includes (as had been the case for each semester) updates which gave some focus to the absolute glut of web resources and repeated announcements given in the lab. Pages on this site which deal with specific topics are still accessible and updated from time to time and will probably all be incorporated into the above-mentioned "General Micro Lab Topics Web Pages." Thus the "Bacteriology 102 Site" is characterized as "semi-retired."
- After using Learn@UW as an on-line course supplement for awhile, I went back to the web with a new MICROBIOLOGY 102 SITE (now on jlindquist.com). Analogous to "Course News" and "Content" on Learn@UW are the easier-to-access updates and links (to course-related matters) on the home page. Through Spring Semester, 2014, this site continued to be as organized and useful as possible to students taking the course. Starting with the Summer Session of 2014 (at the end of which I retired), we returned to the use of Learn@UW as our primary on-line organizer and resource. And with that, this site joined the above-mentioned Bacteriology 102 Site in "semi-retirement." (Note our site outline here.)
|
![]()
![]()
![]()
![]()



 John A. Lindquist. 1991.
John A. Lindquist. 1991.