Differential Media:
|
|
General Principles Applicable to pH-Based Differential MediaIn our introductory microbiology lab course (Microbiology 102), there are several basic topics that we have found advantageous to introduce early in the semester such that we can continue building on them without undue delay. These topics include aseptic technique, dilution theory, elementary catabolism, and differential media – particularly those media which depend on pH-related reactions for their interpretation. The principles that apply to these media constitute the theme of this page and probably could be summarized in a short paragraph. Examples of differential media which are not "pH-based" include Motility Medium, Starch Agar and Thioglycollate Medium, and both kinds of media (pH-based and otherwise) are found throughout this differential media site. The following explanatory table includes hydrogen sulfide (H2S) detection, an additional feature in many of these media that is not pH-related.
How the above table has been summarized in a typical semester of our introductory course is shown below:
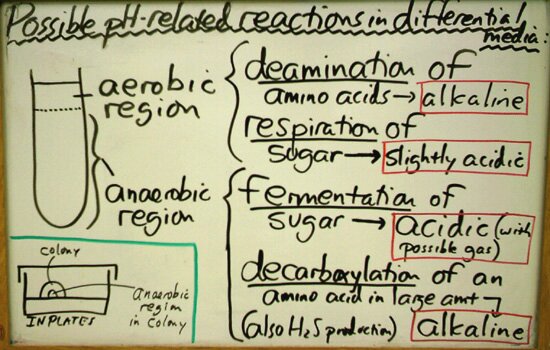
It should be a "given" that most species of bacteria that one works with in an introductory lab will produce an alkaline reaction in media that are formulated with organic compounds. These organic compounds usually include peptones and/or yeast extract which are composed largely of amino acids and peptides – major sources of carbon, nitrogen, sulfur and energy for a wide variety of microorganisms. So, even if a medium is not formulated specifically to be a differential medium whose reactions are based on pH changes, the tendency for many of our organisms while simply utilizing the nutrients is to begin breaking down the amino acids by the process of aerobic deamination which results in an alkaline reaction by the release of ammonium into the medium. The products of amino acid breakdown are utilized in catabolism (usually by respiration) and biosynthesis.
All too often, one may teach or learn that colonies on a plate of MacConkey Agar* are either going to turn the pH indicator (neutral red) a red color due to an acidic reaction (from lactose fermentation) or white due to a neutral reaction (resulting from the lack of fermentation). The alkaline reaction that is there from amino acid deamination often gets ignored by the instructor until it is introduced as a new topic when the time comes to explain such media as KIA or TSI Agar where the interplay between acidic and alkaline reactions becomes critical in the correct analysis of the reactions. So, colonies on MacConkey Agar are going to be inherently alkaline unless the acid from lactose fermentation over-neutralizes the alkaline reaction, resulting in a net acidic reaction. Click here* for one of our "white-board summaries" of MacConkey Agar. In a tube of a medium (such as what is presented in the diagram above) which is inoculated with a pure culture of an organism, various alkaline and acidic products that may be produced by the organism (according to its capabilities) diffuse from their respective origins in the aerobic and anaerobic regions of the tube.
The over-neutralization in the second point can be a problem in a medium where we wish to determine whether or not an organism can ferment glucose. Therefore, the formulation of Glucose Fermentation Broth* allows at least some net acidic reaction to be evident for those organisms that ferment the sugar weakly. A further reduction of peptone is seen in the formulation of Glucose O/F Medium* which is useful in the detection of the very small amount of acid associated with aerobic respiration by such non-fermenters as Pseudomonas. So, if an organism is producing an acidic reaction when tested in (for example) Glucose Fermentation Broth, any net alkaline reaction which may appear in the aerobic region need not be recorded. As long as some acidic reaction is seen in the tube (at least in the anaerobic region), the organism is recorded as a glucose fermenter. Taking advantage of competing acidic and alkaline reactions.
A good example of a useful differential medium in which we can "take advantage" of acidic and alkaline reactions that may over-neutralize each other is Kligler Iron Agar* (KIA) – a medium that is introduced in our course late in the semester when we get to the enteric identification tests. This medium includes glucose in a relatively small amount (0.1%) and lactose in a relatively large amount (1.0%).
So, as might be inferred from the above, a differential medium can be formulated ("programmed") with certain substrates in order to exploit the characteristics of one or more particular physiological types of organisms. The net result from competing alkaline and acid reactions can assist greatly in the detection, differentiation and identification of such organisms. Adding decarboxylation to the mix.Containing a large amount of ornithine in the formulation, a tube of Motility Indole Ornithine Medium* (MIO) can allow the detection of ornithine decarboxylation which will cause a net alkaline reaction in the anaerobic region of the tube. Media used to detect amino acid decarboxylation are generally formulated with glucose to allow anaerobic growth (associated with fermentation) in the first place, and the small amount of glucose would not overneutralize the alkaline reaction from decarboxylation (nor the alkaline reaction from amino acid deamination in the aerobic part of the tube). Consider the search for Salmonella colonies on a selective-differential medium inoculated from a food sample or clinical specimen – and furthermore how colonies of a typical strain of Salmonella appear on the multi-substrate XLD Agar* which includes three fermentable sugars – xylose in a relatively small amount and lactose and sucrose which are each in a relatively large amount; also included is the amino acid lysine which (if decarboxylated) will cause a significant alkaline reaction. So, for a typical strain of Salmonella, formation of the alkaline product from lysine decarboxylation over-neutralizes the small amount of acid produced by xylose fermentation; neither of the other two sugars are fermented by this organism, so a net alkaline reaction results. Also, the production of hydrogen sulfide (a non-pH-related reaction) is shown by the black centers of the colonies. So any alkaline colony with a black center can be considered a possible Salmonella. On most other differential plating media – in which a net pH reaction arises from activity on a smaller variety of substrates – Salmonella may share a colony appearance with a greater number of other organisms, so a search for possible Salmonella colonies (to undergo further testing) can be made a bit more precise by the use of XLD Agar as is also shown here.* The "programming" of differential media.As an example of how a plating medium for isolation can be "programmed," let's start with MacConkey Agar,* a medium which includes lactose as the only fermentable sugar. If we want to use a medium such as this to help us detect and isolate an organism that does not ferment a lot of sugars, one can add a number of these sugars to MacConkey Agar and pick just the non-fermenting (i.e., alkaline) colonies for further study. With this as a starting point, the idea is further developed in the scenario depicted on this page.* Making sense out of this.For a real learning experience from studying the pH-related differential media on this site – most notably the selective-differential plating media and the multipurpose differential media in tubes – see if the net pH result shown for any organism on any of these media makes sense to you. The summary table above can also be applied to other media where pH is the main differential feature. So, rather than approach each medium as a "special case" that has to be analyzed "from scratch," one can apply a pattern or set of principles that can make understanding these media a lot simpler. Such an understanding can help in situations where formulation of a new medium may be required. One can get a general "feel" for this sort of thing on this page.* On a "thought questions" page (on our old Bacteriology 102 website), a few hypothetical situations regarding media formulation are posed with the use of real and/or fictional organisms. Regarding the first two of these questions:
|
|||||||||||||||||||||||||||||||||
|
Why not e-mail me |
These general microbiology pages have copyright by John Lindquist Page content was last modified (and hopefully |
 Clicking on the links
Clicking on the links